Welcome to YY’s personal blog!
Here, you’ll find a collection of in silico codes from my computational and experimental research. Explore tools and insights bridging theory and practice in biology.
View My Posts

About me
I'm a Ph.D. candidate in Human Biology at the University of Tsukuba, advised by Prof. Akiyoshi Fukamizu in biochemistry and molecular biology, and Prof. Yasuteru Shigeta in computational sciences.
My work combines traditional wet lab experiments with bioinformatics, allowing me to explore biology's complexities through both hands-on research and computational analysis. This blend of skills enables me to uncover molecular mechanisms and their health implications, effectively linking laboratory findings with data-driven insights.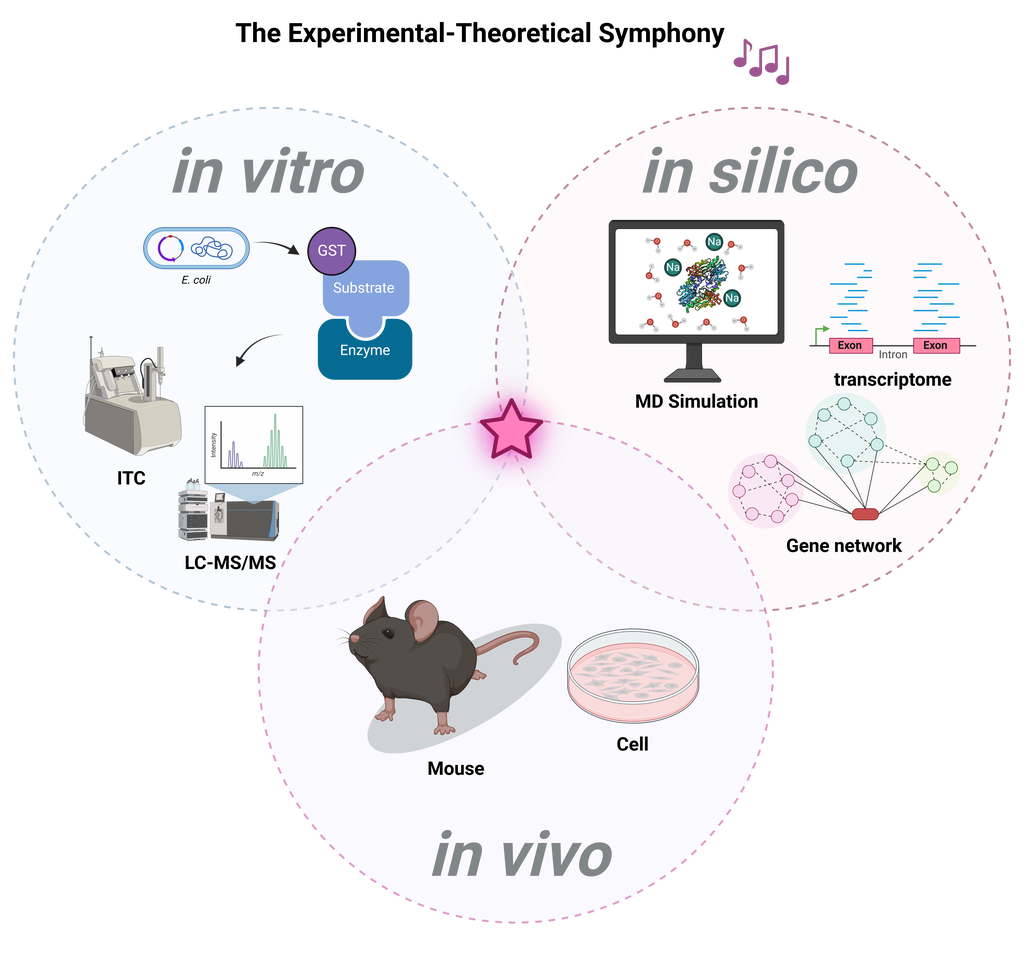
My Research Concept
My research philosophy,The Experimental-Theoretical Symphony, reflects my dedication to integrating experimental biology with theoretical and computational approaches to uncover the complexities of biological systems.
- In Vitro: I measure the activity of externally expressed proteins to understand their functions in a controlled setting.
- In Silico: Through MD simulations, I predict and analyze biological reactions, extending our insights beyond experimental limitations.
- In Vivo: Employing mouse models, I validate theoretical predictions and explore biological processes within a living organism.

My Bioinformatics Research Toolkit
This personal homepage is dedicated to sharing my journey and creations in Bioinformatics, focusing on:
- Employing molecular dynamics (MD) simulations to model protein-protein interactions.
- Developing extensive scripts for molecular docking simulations aimed at high-throughput screening of protein-small molecule interactions.
- Advancing RNA-seq analytical workflows through the creation of automated scripts for quality control, trimming, mapping, and extraction of reads data from large datasets of fastq files.
- Conducting comprehensive downstream RNA-seq analysis using R programming.
- Analyzing RNA-seq data through Weighted Gene Co-expression Network Analysis (WGCNA) to construct co-expressed gene networks for genes of interest.