RNA-Seq
- Home /
- Categories /
- RNA-seq
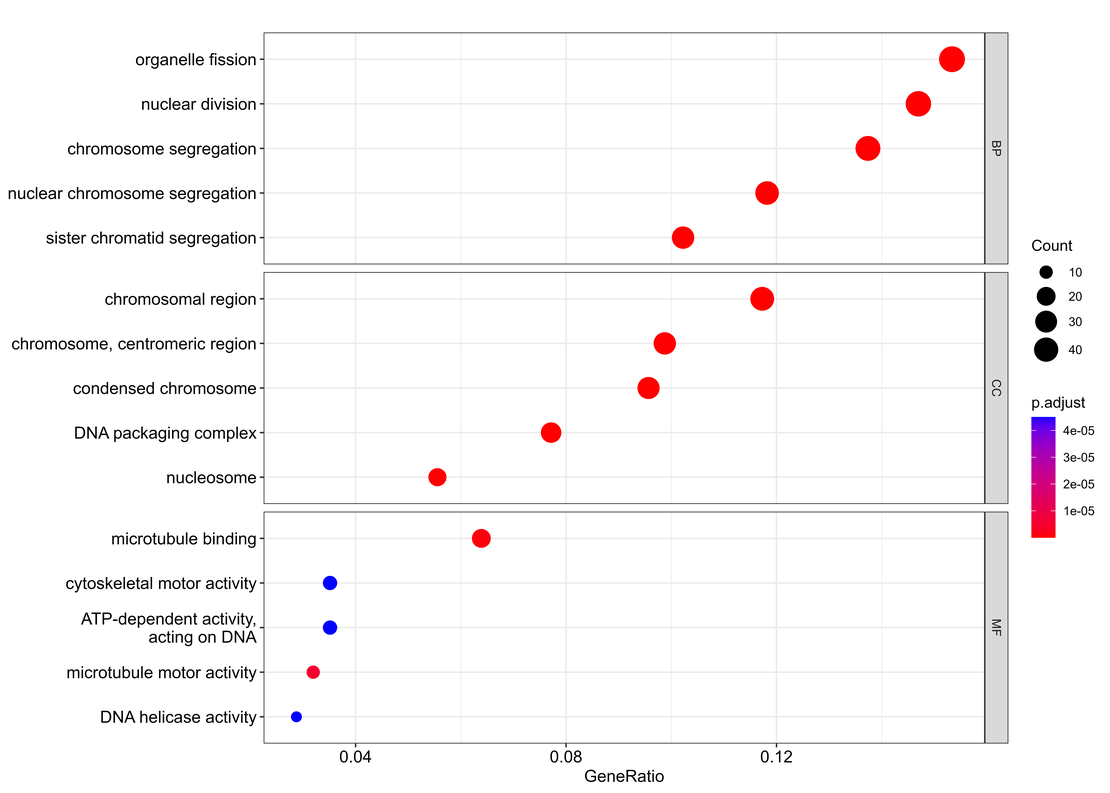
Bulk RNA-seq (9): Gene Ontology Analysis of Differential Expression
Note Next, we will perform Gene Ontology (GO) to unearth the biological processes, cellular components, and molecular functions our significant genes are involved in.
Read More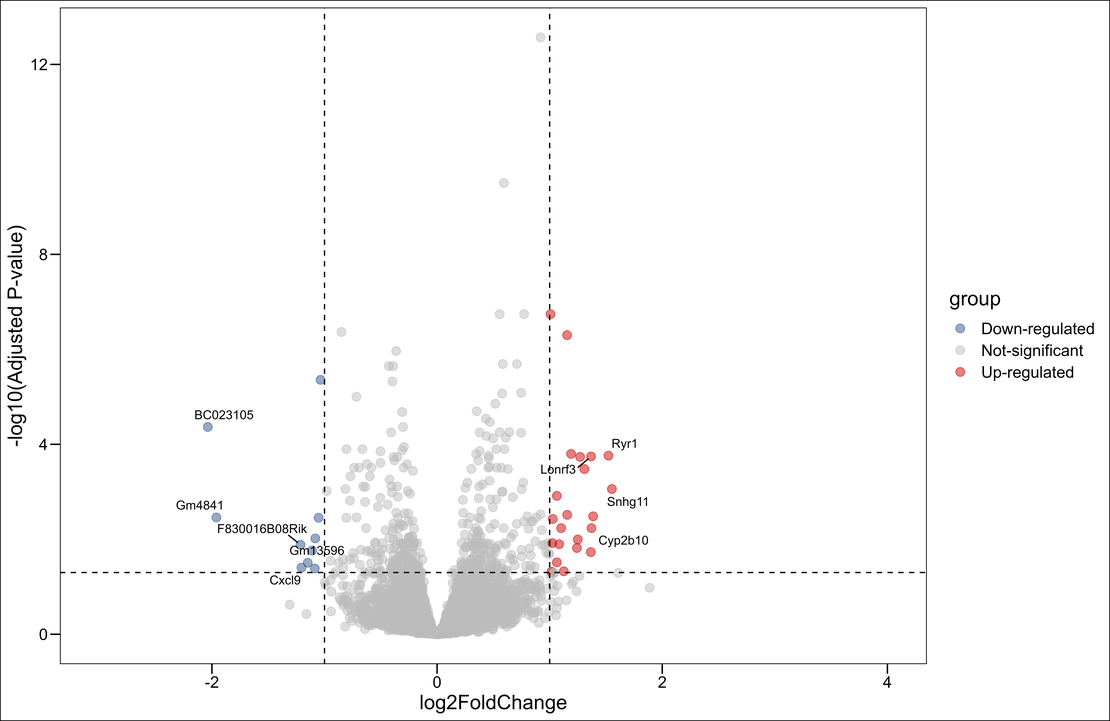
Bulk RNA-seq (8): Differential Analysis and Volcano Plotting with DESeq2
Note Here, we demonstrate how to sift through the complex data to identify genes of interest and showcase their expression patterns through an elegant volcano plot.
Read More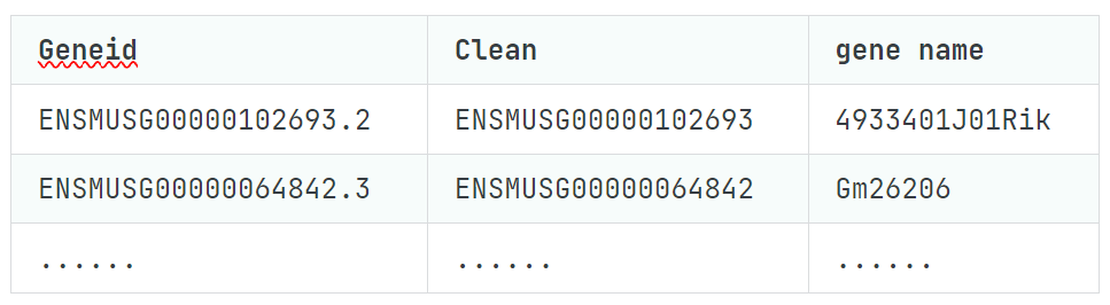
Bulk RNA-seq (7): From ENSEMBL ID Cleanup to Gene Name Conversion
Note In the realm of RNA-seq analysis, one often encounters the challenge of working with ENSEMBL IDs that include decimal points, a result of gene versioning in databases.
Read More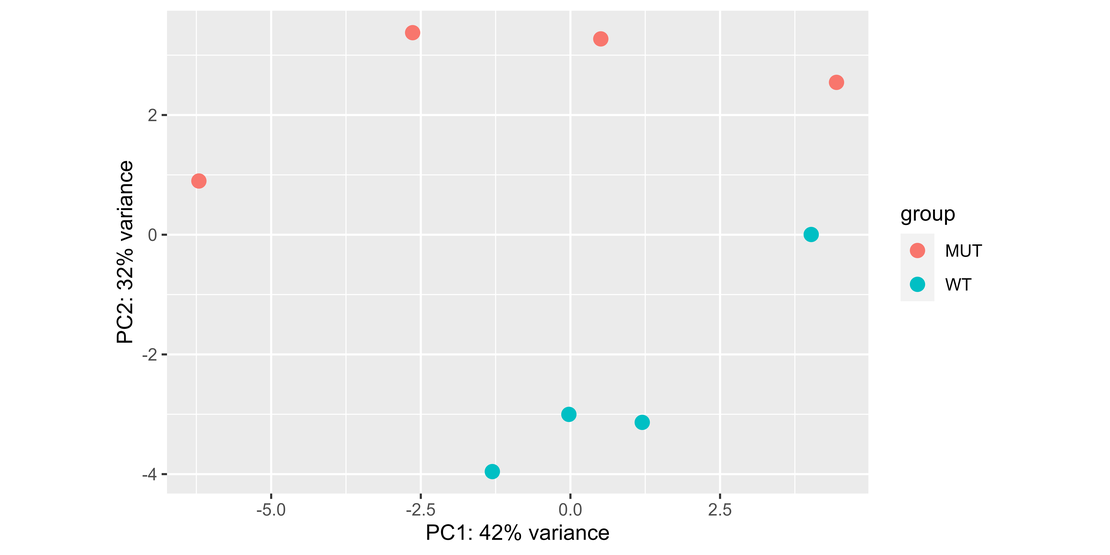
Bulk RNA-seq (6): Normalization and PCA Visualization in RNA-seq Data Analysis with DESeq2
Note In this blog post, we’ll dive into a practical guide on using DESeq2 for differential expression analysis in RNA-seq data.
Read More
Bulk RNA-seq (5): Streamlining Mapping with a Custom Linux Script
Note To overcome the inconvenience of manually mapping each RNA-seq sample to the reference genome, I’ve developed a Linux shell script.
Read More
Bulk RNA-seq (4): Mapping and Quality Control with STAR, Qualimap, and featureCounts
Note This section of the blog series will guide you through mapping RNA-seq read files to a genome index using STAR, performing quality control (QC) with Qualimap, and generating count data with featureCounts.
Read More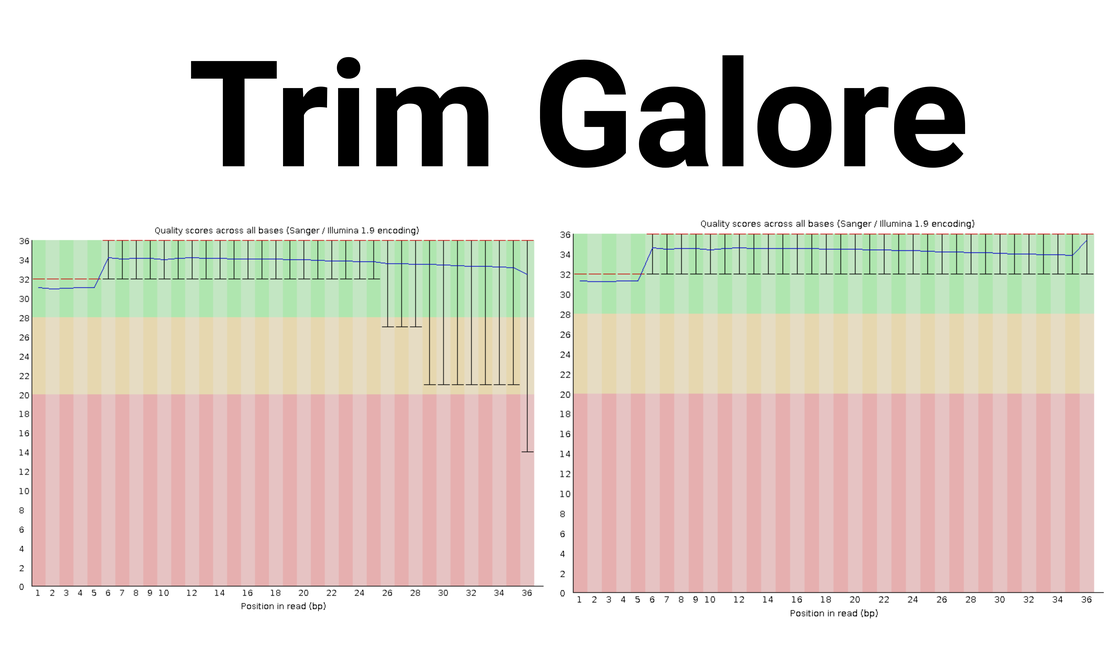
bulk RNA-seq(3):Trim the read files using trim_galore
Tip We will install Trim Galore and trim the merged read files to improve the quality of our data.
Read More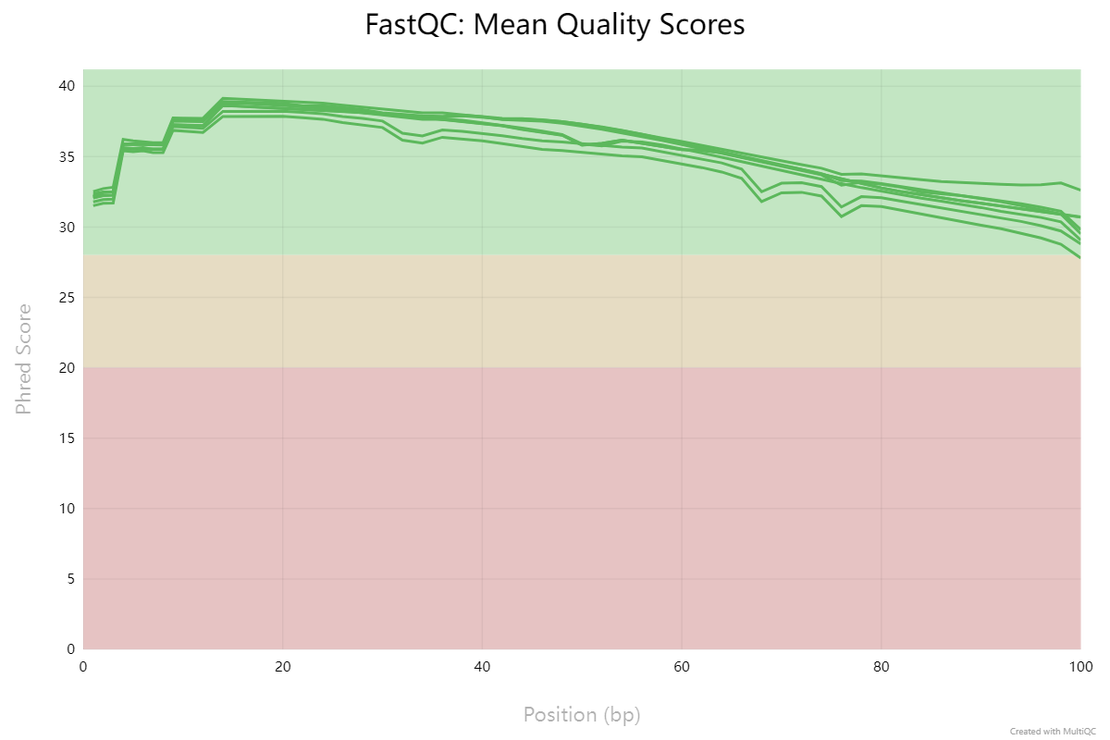
bulk RNA-seq(2):Quality Control with FastQC and MultiQC
FastQC for Quality Checks FastQC provides a simple way to perform quality control checks on raw sequence data.
Read More
bulk RNA-seq(1):Concatenation of raw read files
Concatenation of Raw Read Files To begin with, my operating environment is Linux Ubuntu 20.
Read More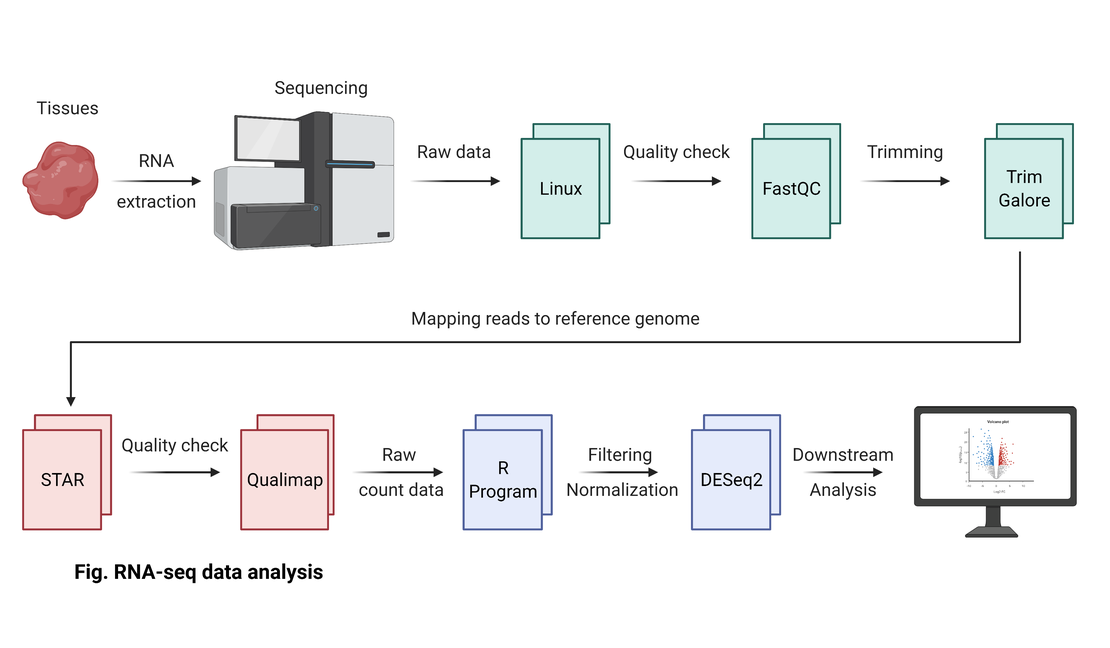
What is the RNA-seq analysis?
Overall description of RNA-Seq The transcriptome has a high degree of complexity including multiple types of coding and non-coding RNA species, like mRNA, pre-mRNA, microRNA, and long ncRNA.
Read More